Project 5
HIV-1 capture and spread at nanoscopic resolution
Summary Project description 2nd funding period
Most studies of HIV-1 infection so far applied ensemble measurements, and few analyses of individual events at high spatial and temporal resolution are available. Cellular and viral macromolecular complexes are involved at all stages, but surprisingly little is known about fundamental details of early replication. Important examples include
(i) attachment and entry,
(ii) timing and mechanism of uncoating,
(iii) structure and composition of the replication complex and (iv) nuclear import of the pre-integration complex (PIC).
Within this project, we focus on image analysis of early HIV-1 replication in different target cells at high spatial resolution applying superresolution (STED) and correlative light and electron microscopy (CLEM). The project is directly connected to the projects Müller regarding dynamics of early events, Lemke regarding nuclear transport of the PIC, Fackler analyzing HIV-1 transmission in complex systems, and Lusic/Beck applying cryo-CLEM to nuclear pores. Furthermore, the project benefits from the central projects on CLEM and chemical biology tools.
In the current period, we have validated a novel experimental system to visualize productive HIV-1 replication complexes in different cell types. This approach distinguishes complexes that have undergone DNA synthesis from non-productive structures. We have also established STED microscopy and CLEM of HIV-1 entry and post-entry complexes with a focus on perinuclear and nuclear events. The cellular protein CPSF6 was identified as a key factor for nuclear import of HIV-1 PICs in primary human macrophages. Nuclear PICs strongly accumulated CPSF6 and a fluorescent version of this protein could be used for CLEM. Knock-down of CPSF6 or a mutation abolishing CPSF6 binding caused accumulation of PICs at nuclear pores as determined by STED microscopy. Importantly, these structures and the nuclear PICs retained (at least some) viral capsid protein (CA) and EM of nuclear PICs suggested a cone-shaped morphology reminiscent of the viral capsid. Based on our results, we propose that consecutive and competitive binding of the nucleoporin Nup153 and of CPSF6 to overlapping binding sites on the viral capsid lattice leads to nuclear import of the HIV-1 PIC and its release from the nuclear basket, at least in post-mitotic primary human macrophages.
In the next funding period, we will continue our investigation of nuclear import of HIV-1 replication complexes and the role of the viral capsid, extending this to T-cells and including spreading infections via virological synapses and in more complex systems (with Fackler). A focus will be on the post-entry capsid and its interactions with host factors applying direct labeling of the incoming capsid by click chemistry (with Müller), copper-independent labeling of nascent DNA (with Johnsson) to preserve ultrastructure and to allow live cell tracking, and cryo-CLEM of HIV-1 PICs at the nuclear pore (with Beck). We will also determine how changes in nuclear import affect selection of integration sites using FISH and next generation sequencing (NGS) (with Lusic), and perform structural and functional analyses on intranuclear PICs. These studies will be done for primary macrophages and extended to CD4+ T-cells. In collaboration with Müller, whose project studies dynamic interactions of the incoming subviral structure with various host factors, we will perform superresolution microscopy and CLEM of potentially relevant interactions with host factors. The role of capsid and its uncoating are currently highly debated, and direct high resolution imaging is likely to help resolve this important question.
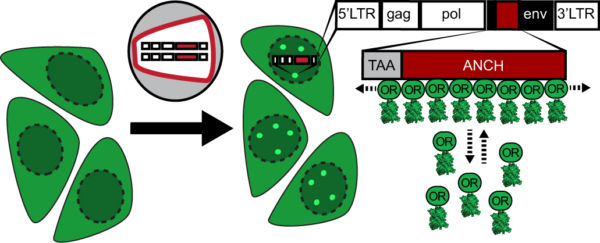
Scheme of the ANCHOR dsDNA visualization system. Fluorescently tagged OR3 binds to the ANCH sequence introduced into the HIV-1 genome.

Imaging of HIV-1 cDNA signal formation in living cells

CLEM-ET renderings of conical and capsid-like HIV-1 complexes inside the nucleus. Internal electron-density indicates the presence of the RNP complex in apparently intact capsids. Tubular and ruptured capsids often appear empty (right panel, bottom capsid). The middle panel shows a positions that correlated to both, fluorescently tagged IN and the OR3 marker, and might show a capsid captured in the process of uncoating.
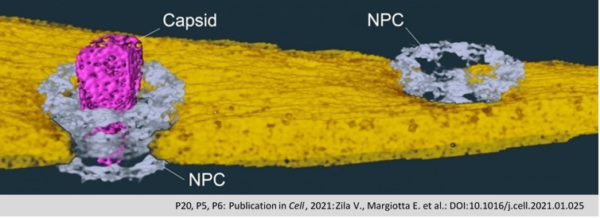
Tomographic reconstruction and isosurface rendering highlighting a conical capsid within a nuclear pore complex. capsid, magenta; nuclear envelope, yellow; NPC, cyan.
The protein capsule of the HIV-1 (Capsid; pink) passes as a whole through the nuclear pore (NPC; grey) into the cell
nucleus, where it disintegrates and releases the viral genetic material. The nuclear membrane is shown in yellow.
(Zila et al., Cell 2021).
.
Project staff
Thorsten Müller, Postdoc
Vojtech Zila, Postdoc